
 |
Manual |
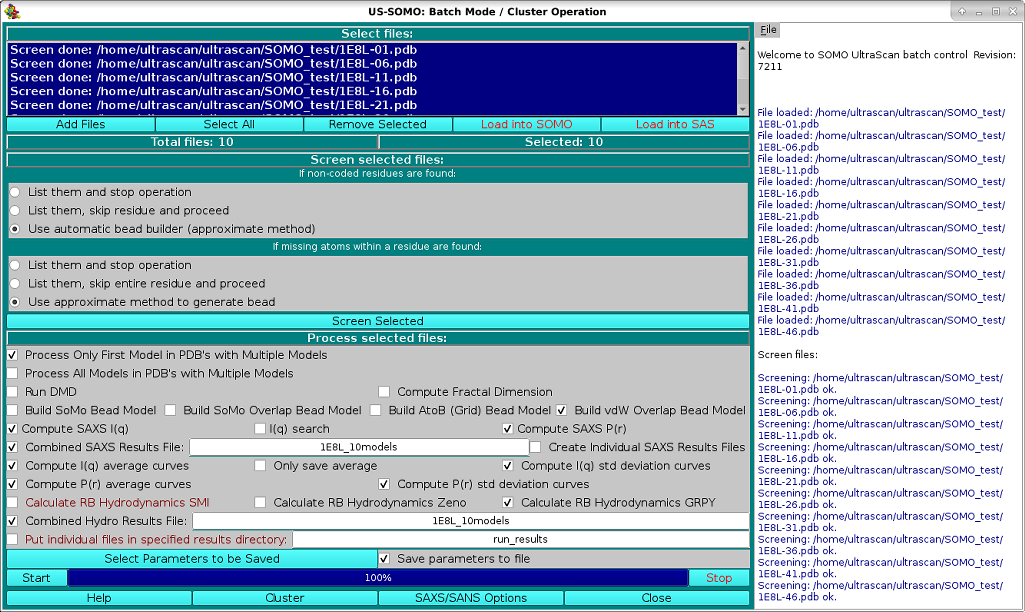
This module has been devised to allow the unattended processing of several files, from the generation of the bead models or of multiple atomic-level conformations via a Discrete Molecular Dynamics (DMD) module, to the computation of several parameters (hydrodynamic, SAXS/SANS).
The operations begin by loading the files in the module via the Add Files button. Several files can be loaded at the same time from the same directory location. Files can be of both kinds supported by US-SOMO, PDB structures or bead models. If unwanted files have been inadvertently loaded, or they are found later to be not suitable for processing (see below), they can be removed from the list by clicking on them and pressing the Remove Selected button. Once the list is completed, a single file can be selected by clicking on it, and multiple files can be selected by clicking on them while holding the shift key. Select All will obviously select all files. The Load into SOMO and Load into SAXS buttons are active when exactly one file is selected and will load the file respectively into the SOMO main window or the SAXS/SANS main window.
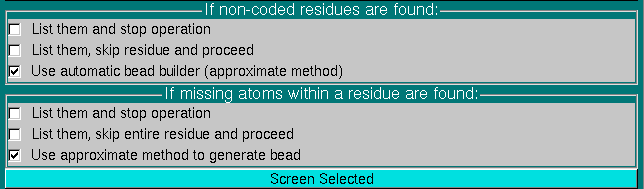
In order to allow for the unattended operations, the files must be checked for compliance with the US-SOMO requirements.
In particular, their residues must be defined in the somo.residue lookup table and must contain all atoms. However, starting from the May 2015 release, the parsing options have been set by default so to cope with incompleted/not coded residues, by the use of approximate methods (a detailed description of these options can be found here). Users can revert to the more stringent List them and stop operation or to the non-recommended List them, skip (entire) residue and proceed options.
For bead models, they must match the overlap tolerance as set in the Hydrodynamic Computations Options module if the Calculate RB Hydrodynamics SMI checkbox is selected (see below).
The uploaded files are therefore screened for compliance with the settings shown in the Screen selected files: panel, by pressing the Screen Selected button. The progress window on the right side reports the progress of the operations, and list the problems that might be found with the files, prompting for corrective action to be taken.
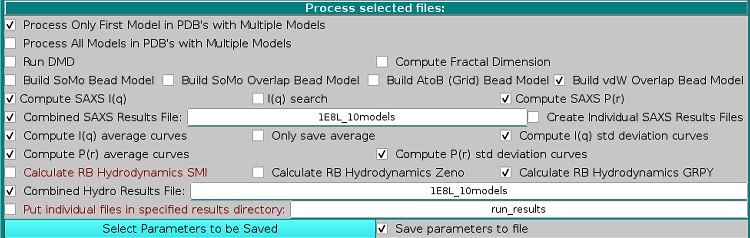
The sequence of batch operation is then set in the Process selected files: panel:
The alternative Process Only First Model in PDBs with Multiple Models and the Process All Models in PDBs with Multiple Models clickboxes control if just the first or all models should be processed in PDB files containing multiple models, like those derived from NMR studies. These options are active only if at least a PDB file is among the selected files.
In new releases produced after January 2023, a temporary directory is automatically created in "...ultrascan/somo/tmp/" (check full path under different operating systems), where the multiple models contained in a NMR-style PDB file will be individually extracted before processing, saving a great deal of computer memory. The advancement of this process in reported in the Progress window on the right side. This temporary directory is then cleared and removed upon batch mode operations completion or halting.
A new feature available since the July 2024 release is the possibility of calculating the Fractal Dimension of structures, by selecting the Compute Fractal Dimension checkbox. This new feature is fully described here.
The Run DMD checkbox will enable Discrete Molecular Dynamics (DMD) runs to be carried on the selected files according to the DMD settings (see here).
The alternative Build SoMo Bead Model, Build SoMo Overlap Bead Model Build AtoB (Grid) Bead Model, and Build vdW Overlap Bead Model dictate which bead modeling procedure will be used to generate the models. These options are active only if at least a PDB file is among the selected files.
Next comes a series of checkboxes relating to SAXS (and in the future SANS) computations (be sure to check the SAXS options currently selected using the Saxs Options button at the bottom of this panel before launching the computations):
Compute SAXS I(q) will allow the computation of the scattering intensity I(q) vs the momentum transfer q curves for each model (see here).
I(q) search is an advanced feature that will allow varying the Buffer electron density, the Scaling excluded volume, and WAT (explicit hydration water molecules) excluded volume in a combined, stepwise manner until a best fit with an experimental curve is found. (see here). IMPORTANT: This selection operates only on local machine computations! For performing an I(q) search on a cluster resource, use the Advanced options feature under the Cluster module.
Compute SAXS P(r) will allow the computation of the distance distribution function P(r) vs r for each model (see here).
Combined SAXS Results File allows the generation of a single csv-formatted file containing all the computed data. A filename should be entered in the field provided.
Create Individual Saxs Results File will also produce individual computed SAXS results files.
Compute I(q) average curves and Compute I(q) std deviation curves become active only if the Combined SAXS Results File checkbox is enabled, and will append to the csv-formatted file extra line(s) containing average and standard deviation values calculated for all the I(q) curves. The Only save average checkbox will enable saving only the average curve.
Compute P(r) average curves and Compute P(r) std deviation curves become active only if the Combined SAXS Results File checkbox is enabled, and will append to the csv-formatted file extra line(s) containing average and standard deviation values calculated for all the P(r) curves.
The alternative Calculate RB Hydrodynamics SMI, Calculate RB Hydrodynamics Zeno, and Calculate RB Hydrodynamics GRPY checkboxes enable the computation of the hydrodynamic parameters after the bead model building phase is completed using one of the four methods available within US-SOMO. If the Build SoMo Overlap Bead Model or the Build vdW Overlap Bead Model checkboxes have been selected above, or if even a single bead model among those selected has the "-so_ovlp" or the "-vdw" suffix in its filename, only the Calculate RB Hydrodynamics Zeno and Calculate RB Hydrodynamics GRPY checkboxes will be available.
Selecting the Combined Hydro Results File: checkbox will produce a single hydrodynamic computations results' file, with at the end the averages of all parameters, instead of separate files for each model uploaded or generated. A filename for the single results' file must be provided in the dedicated space. Otherwise, each file will be named using the general US-SOMO rules and the prefixes present in the main program panel.
A subset of parameters can be saved in another file in a comma-separated format by selecting the Save parameters to file checkbox. The parameters to be saved can be selected from the dedicated module (see here) accessed by pressing the Select Parameters to be Saved button.
To avoid filling up the main SOMO results directory, when the Combined Hydro Results File: checkbox in unselected, the Put individual files in specified results directory:" checkbox can be selected, followed by a (sub)directory name where al the results files will be then placed.

When all the preliminary operations have been completed, the batch operation on your computer can be launched by pressing the Start button, and the progress will be reported in the progress bar and in the right-side progress window. Operations can be aborted at any time by pressing the Stop button. In this case, a warning will appear in the progress window alerting to check for which files are currently still selected in the top panel before relaunching a batch run.
For more demanding computations (and for all the DMD computations), the operations can be transferred to high-end, (super)computing centers using the Cluster button. The SAXS/SANS Options button will instead open the SAS Options panel.
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on June 16, 2024