
 |
Manual |
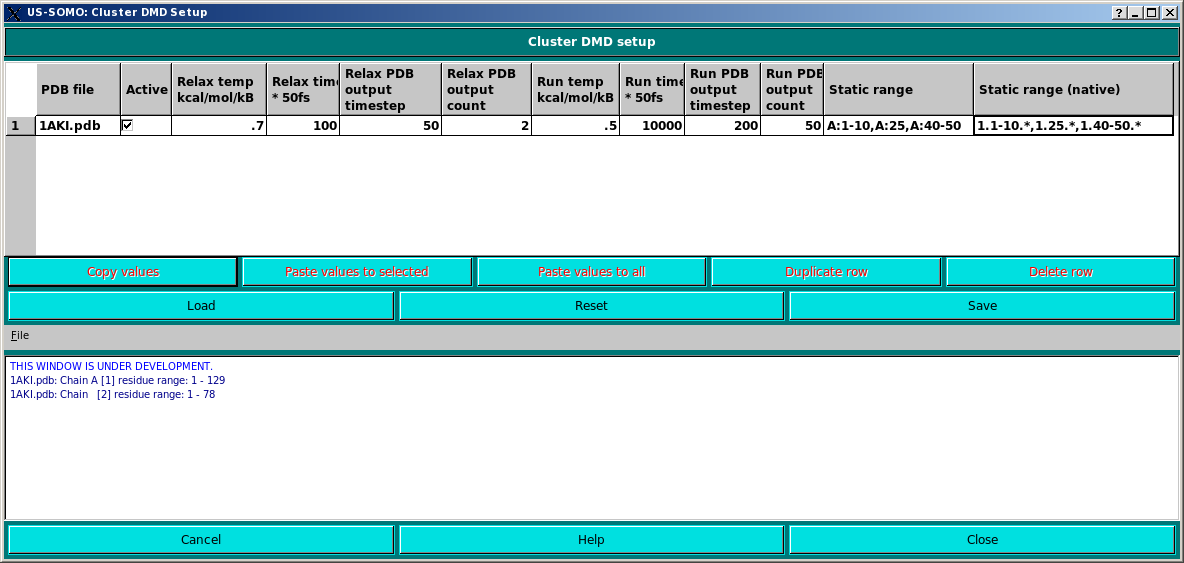
This pop-up windows appears when clicking on the RUN DMD button in the main US-SOMO panel.
The actually operative PDB file is shown in the PDB file field.
N.B.: The PDB should be prepared without any alternate confirmations. Additionally, and missing residue(s) within a chain will create problems and will be broken up by DMD into seperate "chains", due to the impossibility of creating any realistic physical bond across the missing residue(s). Residues not known natively by DMD will cause the run to fail. For example, carbohydrates must be removed. We recommend you run a short run time job until you get a successful run.
Active: whether or not the specific parameters/pdb will be run.
Relax temp kcal/mol/kB: This is the Anderson thermostat temperature for the relax phase of the simulation (default: 0.7 kcal/mol/kB).
Relax time * 50fs: is the duration of the relaxation phase in 50 fs units (default: 200 50 fs).
Relax PDB output timestep: is the time interval for the simulation to output a PDB (default: 100 50 fs).
Relax PDB output count: is the number of PDBs produced (Relax time / PDB timestep = PDB count) (default: 2).
Run temp kcal/mol/kB: This is the Anderson thermostat temperature for the run phase of the simulation (default: 0.5 kcal/mol/kB). Note: multiply by 503 K/(kcal/mol/kB) to get K. To avoid denaturing structured protein regions, run at a cool temperature. Due to the nature of the implicit solvent, it will not freeze.
Run time * 50fs: is the duration of the run phase in 50 fs units (default: 10,000 50 fs). Please do not use longer run times until you have had a successful run.
Run PDB output timestep: is the time interval for the run simulation to output a PDB (default: 1000 50 fs).
Run PDB output count: is the number of PDBs produced (Run time / PDB timestep = PDB count) (default: 100).
The primary difference between the relax and equilibrium phases is the frequency of the heat exchange between the imaginary solvent and the system atoms. The relax phase exchanges heat 100 times more frequently in order to stabilize any energetic stress present in the initial structure. The equilibrium run exchanges heat at a physically realistic rate.
The Static range parameter provides a range of structure parts that will remain fixed during the simulation. This can be entered in a format chain:startresidue#-endresidue#. e.g. A:1-10 will keep residues 1 thru 10 of chain A static. Multiple static ranges can be separated with a comma. N.B.: multiple static ranges will also be static in space with respect to each other.
The Static range (native) parameter is a readonly field that shows the conversion of the entered Static range into the native format used by DMD.
It is possible to select a row by clicking on the row number at the left. This becomes important if you wish to run multiple DMD's simultaneously on the same or different PDB files. Running multiple PDBs simultaneously is actually an efficient use of computational resources, as the DMD program itself is not inherently parallel, and cluster jobs take control of full nodes consisting of from 8 to 16 cores, so each line can run in parallel on the cluster job's allocated nodes.
Copy Values: Copies the values of the selected row.
Paste values to selected: Pastes the previously copied values to the currently selected row. This only becomes available if when Copy Values has previously been pressed.Paste values to all: Pastes the previously copied values to all rows. This only becomes available if when Copy Values has previously been pressed. For example, if you have several PDBs active for a DMD run, you can propagate the various values to all the rows
Duplicate row: This makes a duplicate of the selected row. For example, this could be useful to run the same PDB at various temperatures and/or various static ranges. Running for differing times is not as useful, as the output data of the longer run would just be an extension of the shorter run.
Load: This allows you to load previously saved values. Note: If you have saved valuess for a different PDB, the program will ask if you wish to apply the values to one of the available PDBs.
Reset: Resets to default values and original PDBs. Duplicates of a PDB will be removed.
Save: This allows you to save all current values to a file. It will produce a text file with an extension of .dmd
Cancel: Exits without remembering any changes made to the values.
Help: Brings up this help document in a web browser.
Close: Exits and remembers all changes made to the values in preparation for creating a cluster job package for submission.
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on November 17, 2013.