
 |
Manual |
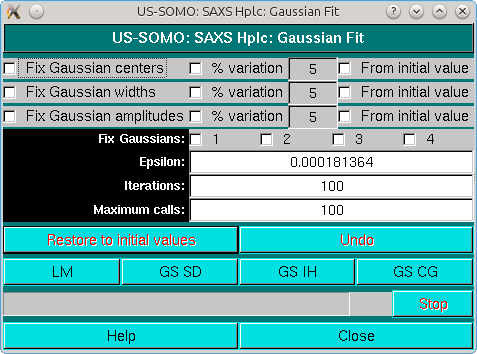
This pop-up panel will appear when the Fit button is pressed in the main HPLC-SAXS module. The panel shown above refers to operations with symmetrical, non-distorted Gaussians. If distorted Gaussians are used, the panel will contain additional fields/checkboxes, as shown below for the EMG+GMG function:
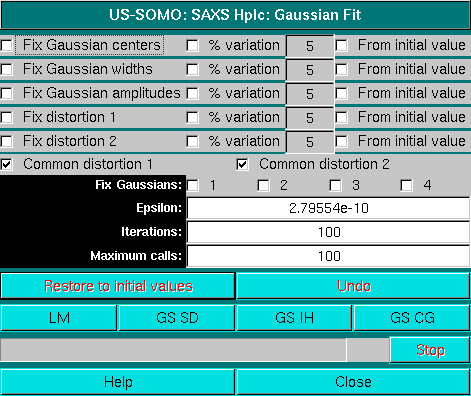
The first three lines in the Fit panel control the centers, widths, and amplitudes of the Gaussians. For each of these parameters, it is possible to fix them to the initial values (checkboxes on the left side), or to allow a % variation (default: 5%) from either the initial values (if the rightmost checkboxes are selected) or based on each cycle of iterations-generated values.
Importantly, since for the most common situation of different aggregation states of similar species eluting from the HPLC column one can safely assume that each peak will present a similar distorsion, by default the Fit module will present one or two (depending on the type of distorted Gaussian function) extra checkboxes called Common distorsion #, already selected. They can be deselected to test the effect of allowing different distorsions for every Gaussian peak. Notice that when you have fixed gaussian(s) and you have Common distortion set, the distortions will only be common to the non-fixed curves.
It is possible to also individually fix each Gaussian by selecting the corresponding checkbox on the Fix Gaussians line; the program will automatically present as many checkboxes as are the input Gaussians.
The Epsilon field controls the step used in computing the discrete derivative, gradient or Jacobian.
In the Iterations field the number of iterations/cycle is set (default: 100).
The Maximum calls controls the attempts to improve at each stage of the minimization.
Several fitting algorithms are available through dedicated buttons: Levenberg-Marquardt (LM), Gradient Search Steepest Descent (GS-SD), Gradient Search Inverse Hessian (GS-IH), and Gradient Search Conjugate Gradient (GS-CG). When initially fitting a single chromatogram, normally a first iteration cycle is performed with the centers fixed, and then a second is sufficient with no constraint or with a restraint on the peak centers from initial values (default 5%) to find a good set of Gaussians.
The resulting Gaussians are updated in the graphics window, and their sum is shown as a dashed yellow line, so that the goodness of the fit can also be graphically assessed. The RMSD of the fit is also updated continuously in its main panel field.
The Restore to initial values and Undo buttons are available to restart the fit procedure from the beginning, or to undo the last operation, respectively.
Close will close the Fit window when a satisfactory fit is obtained
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on May 15, 2014.